





I work on computer vision and medical image analysis, machine learning, and computing pedagogy in the liberal arts environment.
View available software for download.

|
Breast Ultrasound Lesion Synthesis (SPIE-MI '26): Duy Le '27 extended a previously published generative AI model to synthesize tumors in breast ultrasound. He published his work "Tiger-SIREN: Anatomy-Aware Cross-Organ Lesion Synthesis for Breast Ultrasound without Tumour Labels" in SPIE:Medical Imaging 2026 |

|
Facial recognition and machine learning for Bell's palsy management: With Dr. Arun Gadre of Geisinger and Dr. Keith Buffinton of Bucknell, Nick Huo '23 founded our interdisciplary project in facial palsy image processing, receiving a poster award at the regional SVURS'22 (poster, and report). Nick Johnston '24 and Nolan Lwin '26 continued efforts towards a cell phone app that leverages facial recognition software and machine learning to assist in clinical workflow, with Nick receiving a poster award at SVURS'23 (poster). |

|
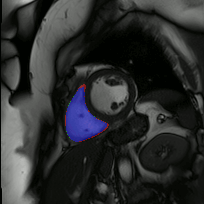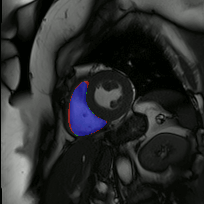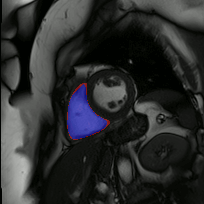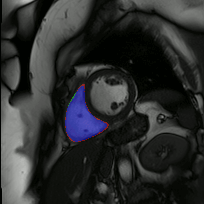
Co-Unet-GAN: a Co-Learning Domain Adaptation Model on Echocardiography Segmentation (SPIE-MI '23): With collaborators at Geisinger Research, Junyang Cai '23 developed a generative adverserial network (GAN)-based deep learning model for domain adaptation across independently-acquired echocardiography datasets. His work shows improved performance over a previously published work. |

|
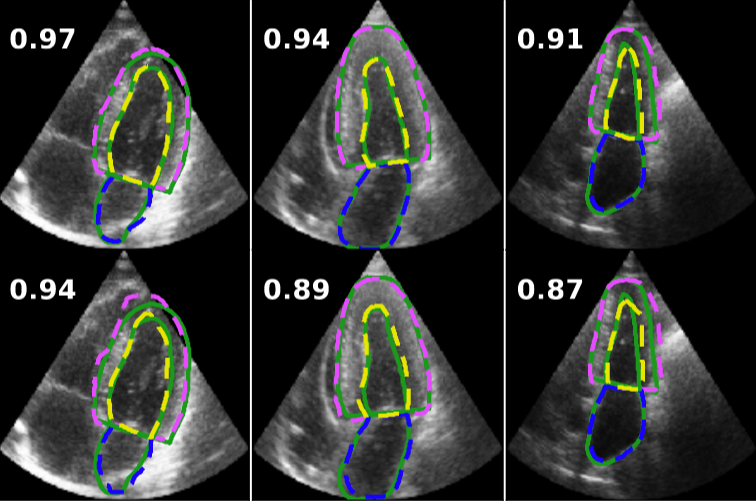
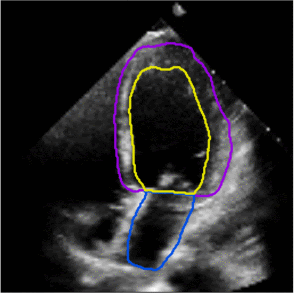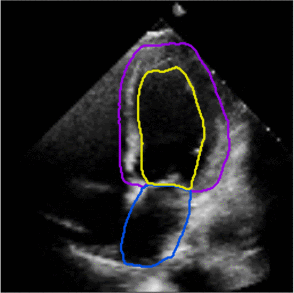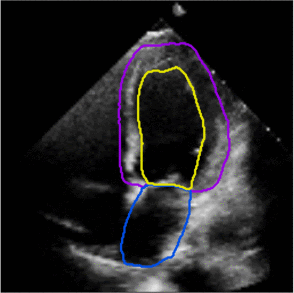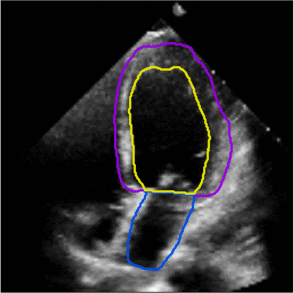
Fully Automated Multi-heartbeat Echocardiography Video Segmentation and Motion Tracking (SPIE-MI '22): In continuing collaboration with Drs. Chris Haggerty and Xiaoyan Zhang of Geisinger Research, Yida Chen '22 extended prior video echocardiography segmentation work, using sliding windows in both training and test time to provide joint semantic segmentation and motion tracking in multi-beat echocardiograms. |

|
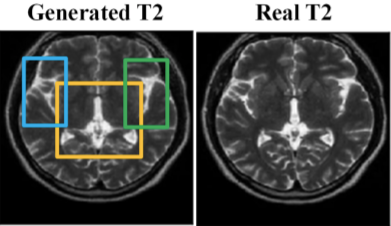
Perceptually Improved T1-T2 MRI Translations Using Conditional Generative Adversarial Networks (SPIE-MI '22): With Dr. Aalpen Patel of Geisinger Research, Anurag Vaidya '21 extended prior image translation work that uses Generative Adversarial Networks (GANs) to estimate a T2-weighted Magnetic Resonance image (MR) given only a T1-weighted MR of the same subject. |

|
Bayesian Optimization of 2D Echocardiography Segmentation (ISBI '21): With Drs. Chris Haggerty and Xiaoyan Zhang of Geisinger Research, Tung Tran '23 applied Bayesian optimization hyperparameter tuning to an established convolutional neural network model for echocardiography segmentation. The resulting optimized model showed dramatic improvement over previous state of the art on a benchmark dataset and this performance generalized to a large independent dataset. |

|
Assessing the Generalizability of Temporally-Coherent Echocardiography Video Segmentation (SPIE-MI '21): With Drs. Chris Haggerty and Xiaoyan Zhang of Geisinger Research, Yida Chen '22 implemented echocardiography video segmentation to ensure consistency between frames, and assessed how such a model performs on a large independent dataset, with comparison to prior work. |

|
Deep Learning for Early Detection of Diabetic Retinopathy (BMES '20): Anurag Vaidya '21 developed binary, early detector, and severity classifiers for diagnosing diabetic retinopathy in fundus images, using convolutional neural networks. His binary classification model provided better predictive capabilities than other state-of-art literature models. Anurag presented his abstract and poster at the annual Biomedical Engineering Society Meeting. |

|
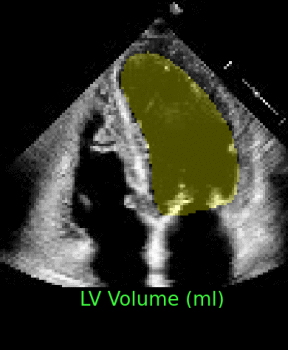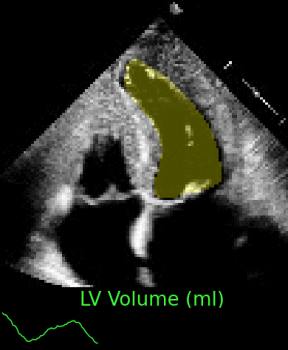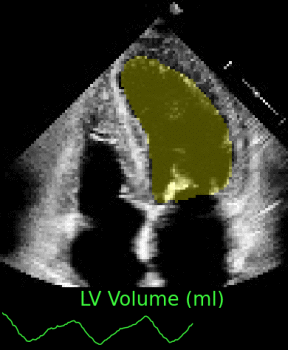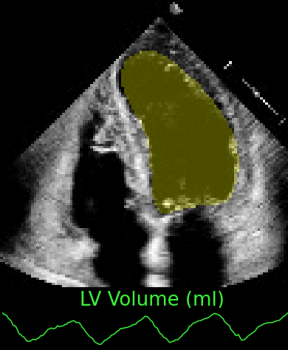
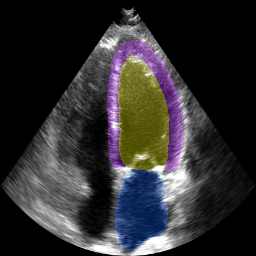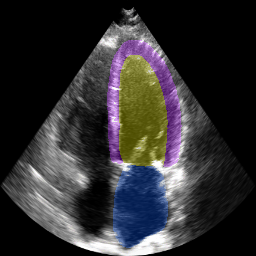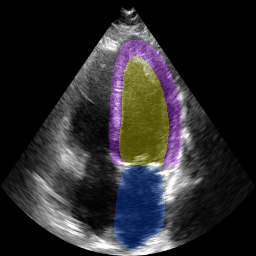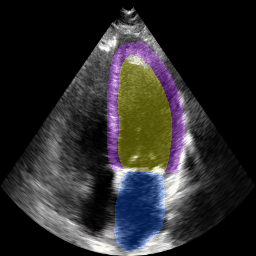
Left ventricular and atrial segmentation of 2D echocardiography with convolutional neural networks (SPIE-MI '20): With Drs. Chris Haggerty and Brandon Fornwalt of Geisinger Research, we developed convolutional neural network learning to automatically segment heart substructures in echocardiography (ultrasound of the heart), acheiving state of the art results in a benchmark dataset. |

|
Ventricular segmentation using convolutional neural networks (SPIE-MI '18): With Drs. Chris Haggerty and Brandon Fornwalt of Geisinger Research, Zilin Ma '19 and Joseph DiPalma '19 used convolutional neural network learning to automatically segment heart substructures in cardiac MR imagery important for understanding cadiovascular disease. |

|
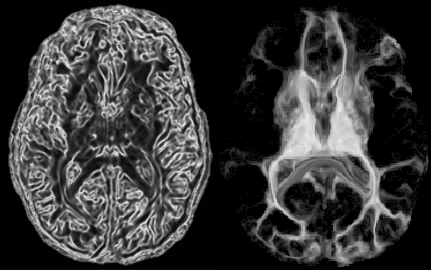
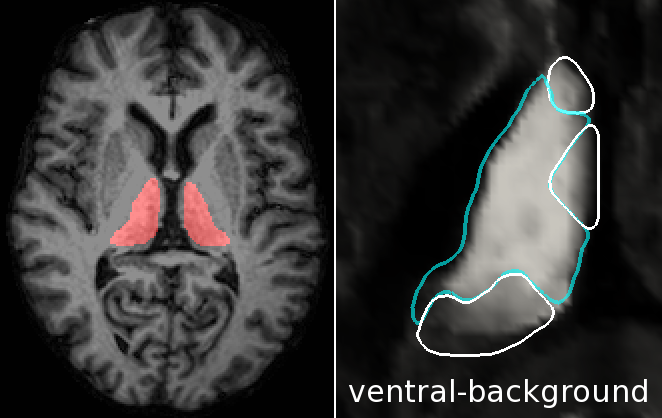
Automatic Method for Thalamus Parcellation Using Multi-Modal Feature Classification (MICCAI '14): In continuing collaboration with Aaron Carass, Jeff Glaister, and Dr. Jerry Prince of Johns Hopkins University, we parcellate the thalamus and its constituent nuclei in a fully automatic way using random forest learning on structural and diffusion-weighted MRI features and computed cortical connectivity. |

|
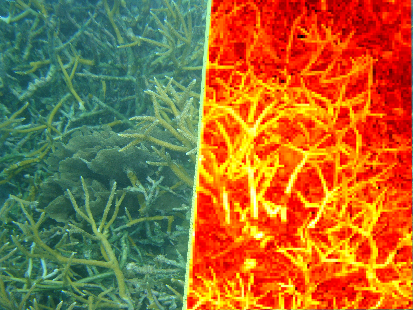
Coral Vision: Software for Improving Efficiency in Coral Monitoring (GSA '13): In continuing collaboration with Dr. Lisa Greer of Geology and her students, Cory Walker '15 created software for the semi-automatic segmentation of coral images. Cory presented her poster at GSA '13 in Denver (see here for more). |

|
Thalamic Nuclear Parcellation Using Multi-Modal MRI (ISBI '13): In collaboration with Chuyang Ye and Dr. Jerry Prince of Johns Hopkins University, we are applying machine learning techniques to segment the human thalamus in magnetic resonance (MR) and diffusion tensor imaging (DTI). The thalamus and its constituent nuclei are differentially affected in neurodegenerative diseases such as mutliple sclerosis and Alzheimer's. |

|
Texture and Color Distribution-Based Classification of Live Coral (ICRS '12): In collaboration with Dr. Lisa Greer of Geology and her students, we are studying automatic abundance estimation of live A. cervicornis with the goal of future rapid assessment monitoring (see here for more). |

|
Condor at W&L (2011): Garrett Koller '14 deployed the Condor distributed computing platform on the CS department's lab machines, with a client-light network-based install. He also developed a Python module to wrap the bash-based submission scripts and intuitively control interaction with the Condor master (see poster, google code project). |

|
DistributedPython (2011): Much distributed computing software is in the end, under many layers, a simple "ssh somewhere doSomething". This code implements that fact in as few layers as practicable. At the top level, you generate a list of command lines and simply request they be executed in parallel (google code project). |

|
Exploiting Parallelism in Computer Vision (2011): Lee Davis '13 and Paul Nguyen '13 explored various parallelizations in natural image object recognition (for the PASCAL competition). Paul implemented and compared OpenMP, Pthreads, and CUDA-parallel MEX implementations of distance matrix calculation for high-dimensional clustering. Lee worked with my DistributedPython module for high-level parallelization across images or training/testing sets (see poster). |

|
Visual Object Recogntion in Natural Images (2010): Will Richardson '11 and Chen Zhong '12 explored improvements to state-of-the-art methods in object recognition for the popular PASCAL competition. Our method used Spatial Pyramid Match and linear Support Vector Machines on densely-sampled SIFT features with the Earth Movers' Distance as a similarity measure for SIFT features. (poster, talk slides for college SSA conference). |
From 2002-2008, I worked in the Medical Image Display & Analysis Group, MIDAG for short, led by Dr. Stephen M. Pizer. During my time in the group, I worked on a number research fronts, mostly appearance models and deformable model segmentation, but also validation and medical image simulation. See the MIDAG bibliography for a complete account of the work that I have been a part of to varying degree.
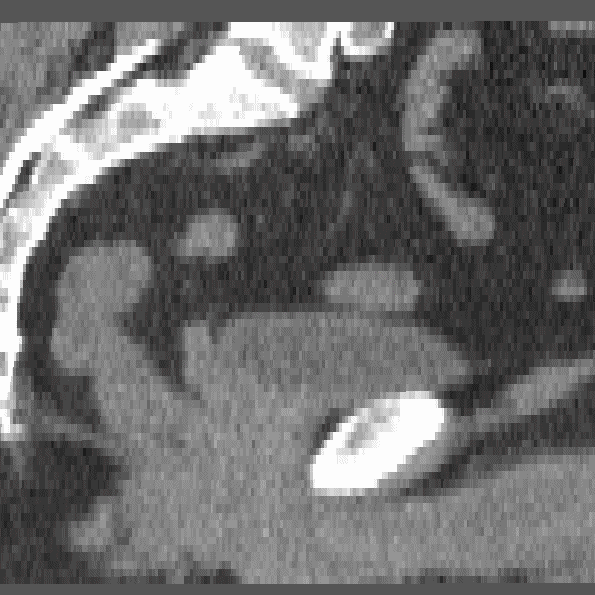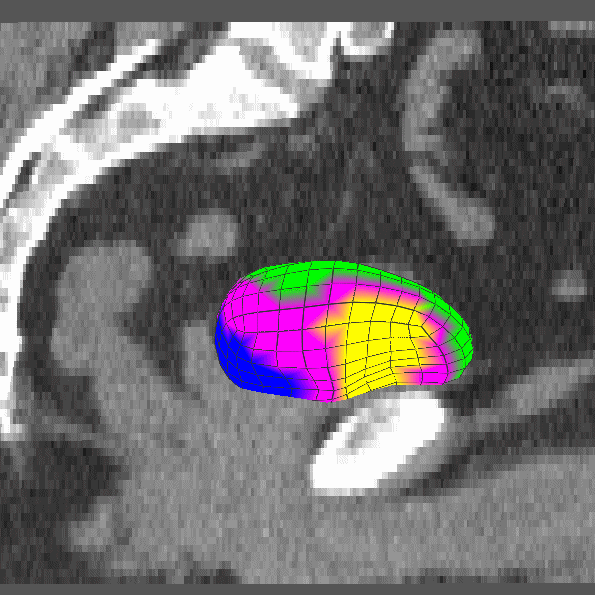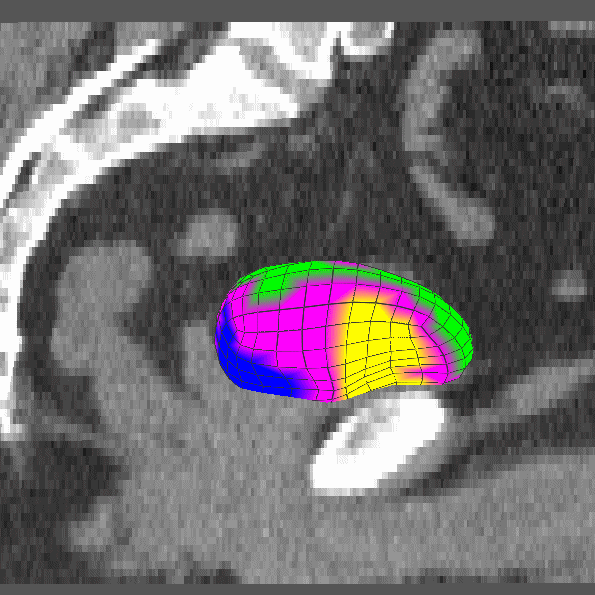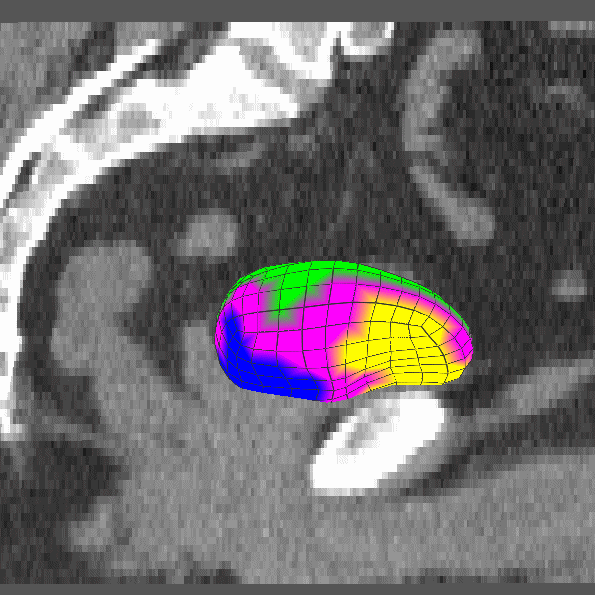
|
Dissertation (2008): Clustering and Shifting of Regional Appearance for Deformable Model Segmentation. |
|
|
IPMI '07: Regional Appearance in Deformable Model Segmentation. |
|
|
ISBI '07: Clustering on Local Appearance for Deformable Model Segmentation. |
|
|
ISBI '04: Clustering on Image Boundary Regions for Deformable Model Segmentation. |
|
|
Compare-byu: a tool for computing volume overlap and surface distance metrics between two surfaces provided as byu object files. |
|
|
Symmetric distance between surfaces: the common closest point distance measure from surface A to B is not the same as that of B to A. I studied diffusion techniques in 3D towards computing a symmetric distance function. |
|
|
Distance quantile functions on bone: we study the pattern of distance from organ to bone to improve prostate segmentation. |