Paper (pdf, preprint)
Slides from the talk
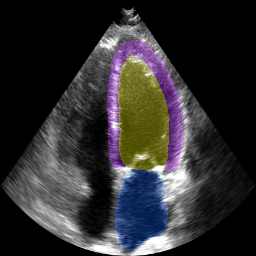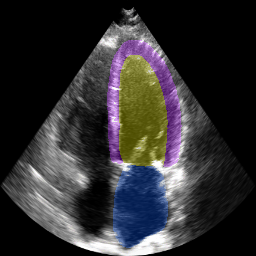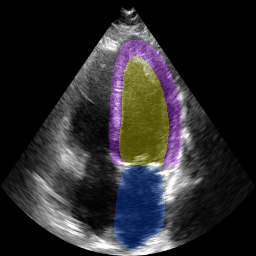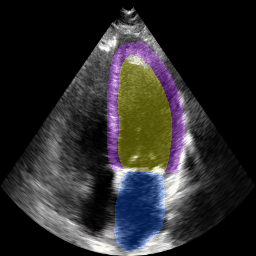
The below animation shows our segmentation results over all time phases for an example apical four chamber echo from the CAMUS datasaet.
Abstract: Segmentation of heart substructures in 2D echocardiography images
is an important step in diagnosis and management of cardiovascular disease. Given
the ubiquity of echocardiography in routine cardiology practice, the time–consuming
nature of manual segmentation, and the high degree of inter-observer variability,
fully automatic segmentation is a goal common to both clinicians and researchers.
The recent publication of the annotated CAMUS dataset will help catalyze these
efforts. In this work we develop and validate against this dataset a deep fully
convolutional neural network architecture for the multi-structure segmentation of
echocardiography, including the left ventricular endocardium and epicardium, and
the left atrium. In ten-fold cross validation with data augmentation, we obtain
mean Dice overlaps of 0.93, 0.95, and 0.89 on the three structures respectively,
representing state of the art on this dataset. We further report small biases and
narrow limits of agreement between the automatic and manual segmentations in
derived clinical indices, including median absolute errors for left ventricular
diastolic (7.3mL) and systolic volumes (4.9mL), and ejection fraction (3.8%),
within previously reported inter-observer variability. These encouraging results
must still be validated against large-scale independent clinical data.
Paper (pdf, preprint)
Slides from the talk
The below animation shows our segmentation results over all time phases for
an example apical four chamber echo from the CAMUS datasaet.
