Abstract: Segmentation of heart substructures in cardiac magnetic resonance (CMR) is an important step in the quantitative assessment of the impact of cardiovascular disease. Manual delineation of these structures, over many patients and multiple time phases, is time consuming and prone to human error and fatigue. In this work we use a deep fully convolutional neural network architecture to automatically segment heart substructures in CMR, achieving state of the art results on a recent benchmark dataset. We further apply our process to a much larger study of CMR subjects, automatically segmenting both left and right ventricular endocardia (LV, RV) with full thirty-phase time resolution, and LV epicardium (Epi) at end-diastole. We validate our automatically obtained results against manual delineations using Dice overlap and Hausdorff distance, as well as Bland-Altman limits of agreement on the derived blood volumes, ejection fraction, and LV mass. We obtain median Dice overlaps of 0.97, 0.94, and 0.97 on the three structures respectively, and further find small biases and narrow limits of agreement between the two assessments (manual, automatic) of volumes and mass. Our results show promise for the fully automated analysis of the CMR data stream in the near future.
Paper (pdf, preprint)
Zilin Ma's Engineering Symposium Poster
Joseph DiPalma's Engineering Symposium Poster
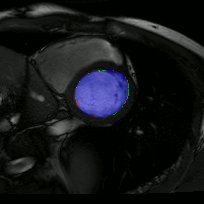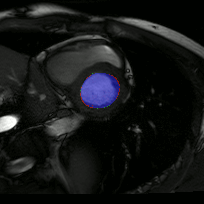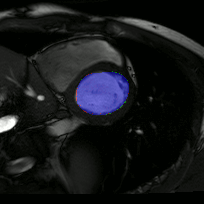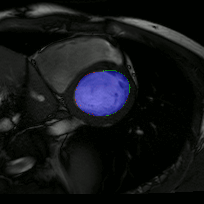
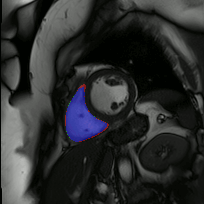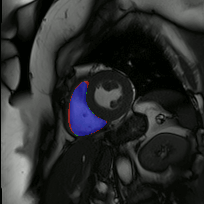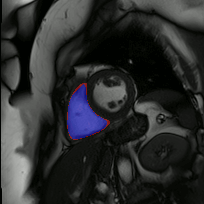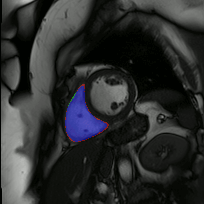
The below animations show our segmentation results over all time phases for example left and right ventricles. Green represents automatic results (false positives), red manual (false negatives), and blue the intersection (true positives).